Junjie Huang1*,Yanchao Xu2, Yunfan Xue1, Peng Zhang1, Junbo Zhao2, Jian Ji1,3*
1MOE Key Laboratory of Macromolecular Synthesis and Functionalization, Department of Polymer Science and Engineering, Zhejiang University, 38 Zheda Road, Hangzhou, 310027, P.R.China.
2College of Computer Science and Technology, Zhejiang University, 38 Zheda Road,Hangzhou, 310027, P.R. China.
3International Research Center for X Polymers, International Campus, Zhejiang University, 718 East Haizhou Road, Raining, Zhejiang, 314400, P. R. China.
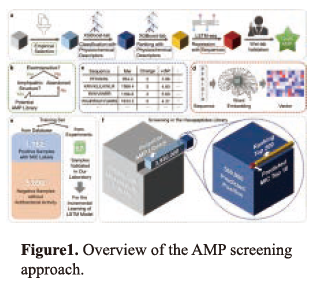
EXTENDED ABSTRACT: The vast combinatorial space of peptide sequences constitutes a huge reservoir for potential therapeutics. However, how to systematically identify the functional peptide sequences from the gigantic search space remains an unsolved research challenge. Here, we report an efficient AI-based framework, named sequential model ensemble pipeline (SMEP). We verify the validity of SMEP on the mining of antimicrobial peptides (AMPs) from the entire hexapeptide virtual libraries that contain 64 million of sequences in 9 d mere. Firstly, in order to avoid the interference of a large number of non-antibacterial peptides in candidates and reduce the computational pressure of the machine learning models, we set up two terminal conditions for peptide screening. 3,930,000 peptides are acquired in this step. Then, they are classified (antibacterial or nonantibacterial) by a classification model with precision of 93% and 560,000 peptides are predicted positive. Thereafter, 560,000 peptides are ranked by their antibacterial ability via a ranking model and the top-500 peptides are chosen. Considering the data noise of labeled AMP we randomly synthesize 67 AMPs and measure their MIC in our laboratory for model modification viaincremental learning. Finally, the MICs of the top-500 peptides are predictedby the modified regression model and the top-10 peptides are selected for wetexperiment verification, It is indicated that all 10 peptides are antibacterial with MIC< 160 μg/μL. Especially, the MICs of 3 best peptides (CRRI hexaAMPs) are lower than 10 μg/uL, better than that of the best antibacterial hexapeptide MP196 (32μg/mL). What' more, the pipeline is used to identify top AMPs from hepta- (1.28 billion of sequences, 13 d), octa- (25.6 billion of sequences, 22 d) and nona- (512 billion of sequences, 27d) peptidevirtual libraries, without changing any parameter. Wet experiment reveals that 98.2% predicted peptides are AMPs (54/55). We further investigate the biological function of the 3 leading CRRI hexaAMPs. In vitro results indicate the bactericidal broad-spectrum, low off-target toxicity, mechanism of lysing cytomembrane, low microbial resistance of CRRI hexa-AMPs. While in vivo results reveal the excellent therapeutic effect to mice acute and chronic pneumonia (sterilizing rate of 99.8% and 98.1 %, respectively). The results above demonstrate the powerful ability of SMEP for AMP identification. This work was accepted by Nature Biomedical Engineering.
Keywords:AMP, machine learning, pipelin
Junjie Huang has completed his PhD from Zhejiang University. Now, he is the Postdoctoral in Zhejiang University. He has long been engaged in the field of machine learning accelerating material discovery and high-throughput experiment accelerating material discovery. He has published 7 papers in journals like Nature Biomedical Engineering (accepted), ACS Applied Materials & Interfaces and so on.